Prevalence, Distribution and Serotyping of Dengue Virus and Characterization of the Non-Structural Gene from Pakistani Isolates
Muhammad Bilal Khan1,2, Deeba Amraiz2, Rehana Rani3, Yeou-Lih Huang1, Hans-Uwe Dahms4,5,6,7*, Liaqat Ali2*
1Department of Medical Laboratory Science and Biotechnology, Kaohsiung Medical University, Kaohsiung, Taiwan.
2Department of Biological Sciences, National University of Medical Sciences (NUMS), Rawalpindi, Pakistan.
3Department of Life Sciences, Abasyn University Islamabad Campus, Pakistan
4Department of Biomedical Science and Environmental Biology, Kaohsiung Medical University, Kaohsiung, Taiwan.
5Research Center for Precision Environmental Medicine, KMU - Kaohsiung Medical University, Kaohsiung 80708, Taiwan, R.O.C.
6Department of Marine Biotechnology and Resources, National Sun Yat-sen University, Kaohsiung 80424, Taiwan, R.O.C.
7Department of Environmental Science and Engineering, College of Engineering, National Pingtung University of Science and Technology, Pingtung 91201, Taiwan, R.O.C.
Correspondence to: Dr. Liaqat Ali, Assistant Professor, Head, Section of Microbiology and Immunology, Department of Biological Sciences, National University of Medical Sciences (NUMS), Rawalpindi, Pakistan. E-mail: liaqatbiotech@gmail.com
Dr. Hans-Uwe Dahms, Professor, Department of Biomedical Science and Environmental Biology, Kaohsiung Medical University, Kaohsiung, Taiwan. E-mail: hudahms11@gmail.com
Received: April 04, 2024; Accepted: April 18, 2024; Published: April 26, 2024
Citation: Khan MB, Amraiz D, Rani R, Huang YL, Dahms HU, Ali L. Prevalence, Distribution and Serotyping of Dengue Virus and Characterization of the Non-Structural Gene from Pakistani Isolates. Arch Virol Infect Dis. 2024;2(1):01-16. DOI: 10.5281/avid.11202417
Copyright: © 2024 Ali L, Dahms HU. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Dengue fever is a mosquito-borne arboviral disease caused by any of four related dengue viruses: dengue virus 1, 2, 3, and 4 (DENV-1-4). The dengue viruses cause millions of infections worldwide every year.
The aim of this study is to identify the identity, prevalence, and distribution of Dengue serotypes during the COVID-19 pandemic (2020-2022) in Pakistan, and to characterize the non-structural gene 3 (NS3) for further functional analysis to make novel therapeutics for dengue viral infections. Dengue virus symptomatic samples were collected, analyzed by ICT method for DENV NS1 antigen and then subjected to PCR to determine the serotype responsible for infection. The identity of the DENV responsible for infection was confirmed through amplification of the DENV serotype-2 and DENV NS3 gene. We found that DENV-2 was the most prevalent DENV in Pakistan during the COVID-19 pandemic. Pakistani DENV-2 sequences were closely related to DENV-2 sequences reported from India, China, Malaysia, Singapore, US and Brazil with homologies above 99%. Our results also showed that males were more affected than females, and we also found co-infections of serotype-2 patients with serotype-3. The occurrence of co-infection with different genotypes or serotypes may raise the disease risk and severity in subsequent outbreaks in Pakistan. These results highlight the need for further surveillance of circulating dengue viruses to aid early diagnosis, determine disease burden and distribution and provide the basis for targeted therapeutics and therapies against dengue infection.
Keywords: Dengue; Serotype; Co-infection; Pathogenesis; Mosquito-borne; NS3; Therapeutic.
INTRODUCTION
Dengue viruses affect mostly tropical regions, where 3.9 billion people live worldwide. Dengue viruses are transmitted to humans through Aedes mosquitoes, mostly by Aedes aegypti and second most by Aedes albopictus. Aedes aegypti is a day-biting mosquito that breeds in stagnant waters and is found in tropical and subtropical areas [1]. Different tropical and subtropical regions have a major public health challenge due to the dengue virus. Primary infection caused by dengue virus results in mild fever, while dengue hemorrhagic fever (DHF), also causes another severe disease, dengue shock syndrome (DSS). both diseases are life-threatening [2].
The Dengue virus belongs to the family Flaviviridae [3,4]. The four closely related viruses that cause dengue infections are DEN-1, DEN-2, DEN-3, and DEN-4. These four viruses are referred to as serotypes since they interact differently with the antibodies found in serum from human blood. The classification of its serotypes is based on their antigens on the virus's surface termed a serotype. The scientists who firstly noticed affirmation of dengue virus strain variations and independently isolated different strains of the dengue virus was Albert B. Sabin and Walter Schlesinger in 1944 [5]. These serotypes are antigenically distinct but closely related and have sixty-five percent to seventy percent sequence homology [6,7]. Each serotype has a unique genotype with extensive genetic diversity, making vaccine development against all four dengue serotypes a challenging task [8]. In Pakistan, four dengue serotypes are prevalent and circulating throughout the year, with a peak epidemic wave of outbreaks between September and November during the post-monsoon season [9,10].
Numerous clinical symptoms are caused by the four serotypes, and the order or timing of infections may have an impact on the severity and prognosis of the disease [11]. Dengue fever can be classified as mild, moderate, or severe, depending on the response to the disease by the response of the patient [12]. The three phases of dengue fever disease include the acute febrile phase where the fever typically lasts for two to seven days and is frequently accompanied by headache, myalgia, arthralgia, facial flushing, and skin erythema. Certain patients may experience injections to the pharynx, conjunctiva, and sore throat. Vomiting, nausea, and anorexia are frequent. In the early febrile phase, clinically differentiating dengue from non-dengue febrile diseases can be challenging. The second phase is a critical phase when the temperature falls to 37.5–38°C or lower and remains at the same level, which typically happens on days 3–7 of illness, there may be a simultaneous rise in capillary permeability and an increase in hemoglobin levels. Clinically significant plasma leakage typically lasts between 24 and 48 hours. Generally, progressive leukopenia precedes plasma leakage and is followed by a sharp drop in platelet counts. When a critical volume of plasma is lost due to leakage, patients may experience a shock. If the patient survives for the following 48–72-hour period, extravascular compartment fluid is gradually reabsorbed if the patient makes it through the critical 24-48-hour phase. Diuresis follows, overall health improves, appetite returns, gastrointestinal symptoms subside, and the hemodynamic status stabilizes [13]. The 2009 WHO categorization replaces the previous WHO approach from 1997, which addressed and emphasized the disease's two clinical manifestations: plasma leakage and disturbed hemostasis. Patients were classified as showing DF which is dengue fever or severe dengue hemorrhagic and shock syndrome (DHF/DSS), i.e. leakage of plasma and coagulopathy, often characterized by bleeding, can cause a fast drop in systolic and diastolic levels, resulting in cardiovascular shock and subsequent damage of organs [14].
Dengue fever comes along with rashes, vomiting, severe headache, nausea, muscle and joint pain, but DHF is defined by a high temperature, hemorrhagic manifestations, hepatomegaly, and often circulatory instability and shock [15]. When people with asthma, diabetes, or other chronic illnesses encounter dengue fever, the infection can be fatal [16].
Regarding the life cycle, there are different types of receptors that mediate the attachment and entry of the virus to host cells. In the early stage, virus particles are attached to receptors and enter via the clathrin-mediated endocytosis pathway. When they enter, they merge with endosomal membranes of cell organelles and are then released to the cytoplasm. The viral protein replicates into a single polyprotein, and after that, it divides into seven non-structural and three structural proteins. The viral assembly takes place in the endoplasmic reticulum, and the viral particles become mature in Trans-Golgi complexes. The mature viral particles are released through exocytosis, which then infect other cells and continues the viral life cycles. A general life cycle of the dengue virus (Figure 1).
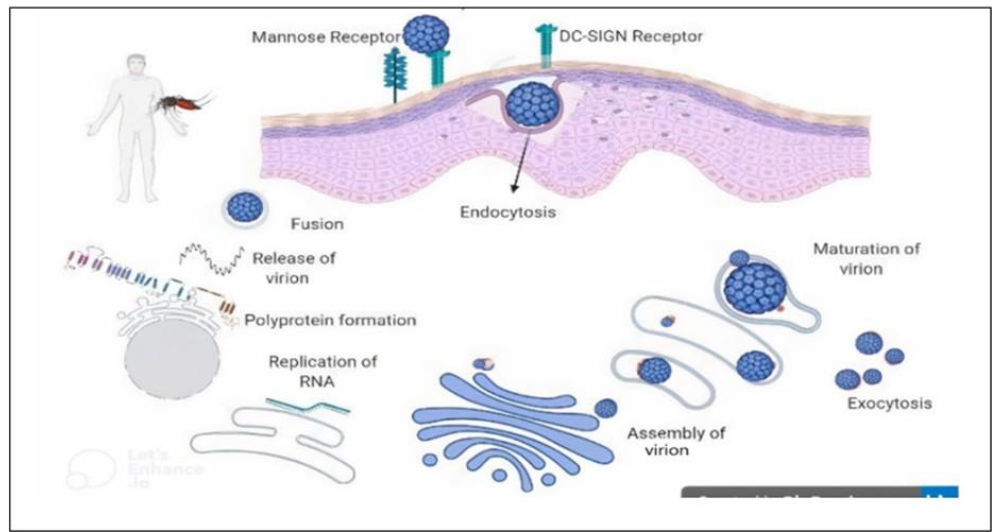
Figure 1: Life cycle of dengue virus (generated by Biorender software, https://www.biorender.com)
A single-stranded positive-sense RNA, approximately 11 kb in length makes up the DENV genome [17,18]. Seven non-structural (NS) proteins, including NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, are encoded by it, along with three structural proteins, including capsid (C), membrane protein (prM/M), and envelope (E), (Figure 2) [18-20].
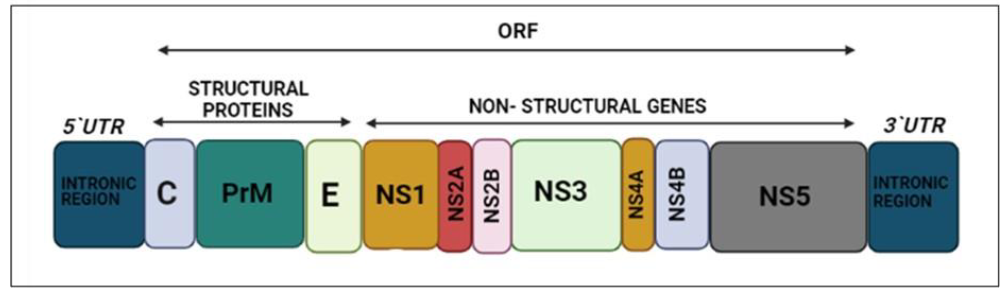
Figure 2: Genomic organization of the Dengue virus (using Biorender, https://www.biorender.com)
Dengue causes ~390 million infections annually worldwide. Dengue cases have increased 30-fold in the past five years [9]. Currently, DENV is endemic in 128 countries, where 70% of the disease cases occur in Asia [9]. Dengue virus was firstly introduced to Pakistan at Karachi Seaport through the import of tires containing eggs of infected mosquitoes in 1994 [9].
Several dengue virus outbreaks occurred in Pakistan in the past three decades. From 1994 to 2016, there have been 71,649 recorded dengue cases in Pakistan and 797 deaths [21]. Dengue outbreaks occur in Pakistan during the post-monsoon season. Dengue virus antibodies were initially detected in 1982 from serum samples obtained in the Punjab province in 1968 and 1978 [1]. Dengue fever shows symptoms like severe headache, muscle and joint pain, rash, nausea and vomiting while symptoms such as high temperature, megalohepatia, hemorrhagic phenomena, shock, and often cardiovascular disturbance are shown by Dengue hemorrhagic fever (DHF) [1].
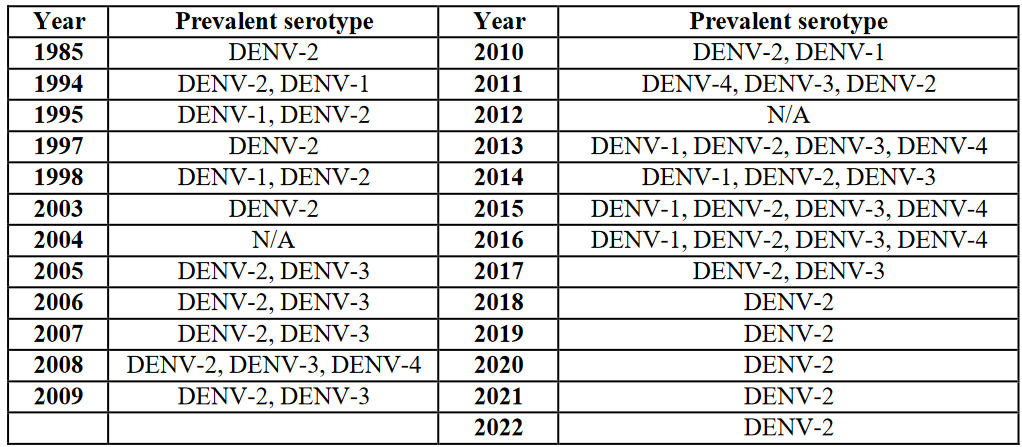
Table 1: Prevalent DENVs in Pakistan from 1985 to 2022 [1, 4, 9, 10, 18, 20-26].
In Pakistan, climate, mosquito populations, and public health initiatives are major variables that have affected the prevalence of dengue virus from year to year [27]. Different serotypes were prevalent from 1985 to 2021; serotype 2 was prevalent every year in addition to other serotypes being provided (Table 1).
Upon first meeting any of the four dengue virus serotypes, a primary infection occurs. High titers of immunoglobulin M (IgM) and immunoglobulin G (IgG) antibodies appear 3-5 to 6-10 days after the start of infection, respectively. After the symptoms are shown in 3-5 days for a few months’ IgM is present, but IgG is persistent throughout one's life [28]. It is not clear yet how immune responses protect DENV's infection or lead to the acute outbreak of disease. When antibodies attach to virus particles in large enough amounts, infection gets minimized and suppressed. As a result, lifetime immunity develops against the same serotype. But cross-protection against the remaining serotypes is not provided (Figure 3) [29].
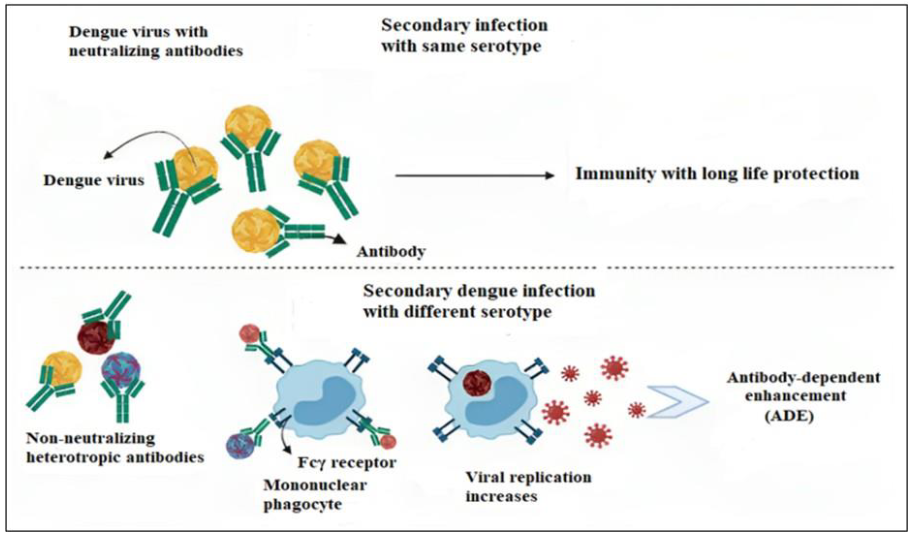
Figure 3: Antibody dependent enhancement (ADE) mechanism. Primary DENV infection produces a long-lasting protective immunity that can neutralize other DENVs of the same serotype (upper part). When a secondary heterotypic DENV infection occurs, anti-DENV antibodies cross-react with the heterotypic DENVs to produce an immune complex. Fcγ receptors are primarily present on phagocytes and macrophages and interact with the virus-antibody immunological complex. The heterotypic DENVs which multiply inside infected immune cells called Fcγ, increase the viral infectivity (lower part) M & M(generated by Biorender software, Toronto, Canada).
In this study, we characterized the different serotypes of DENVs genetically to identify the prevalence, distribution, and predominantly circulating DENVs during the COVID 19 pandemic from 2020-2022. We then amplified the NS3 gene for further functional analysis like cloning and mammalian cell line expression and to check the inhibition of NS3 gene against RNAi for the development of novel therapeutics against the Dengue virus.
MATERIAL AND METHODS
Sample Collection
A total of 267 samples were taken from the individuals having dengue symptoms which includes high fever, nausea, vomiting, muscle and joints pain and severe headache, from different hospitals in Khyber Pakhtunkhwa province and from Islamabad, Pakistan. Of these 267 blood samples, 156 samples were from adult males and 111 were from adult females. Serum was extracted and stored at -80°C.
Immunochromatographic Assay
All the samples were checked and analyzed by the ICT method for dengue NS1 antigen by Bioline™ dengue NS1 Ag ICT kit, (Abbott, ME, USA). Positive dengue samples were used as a control to check the test device.
RNA Extraction and Quantification
Following the manufacturer's instructions, DENV RNA was extracted from serum samples using the Nucleospin viral RNA extraction kit (MACHEREY-NAGEL GmbH & Co. KG). A Nanodrop spectrophotometer was used to measure the quality and quantity of RNA (Multiskan Sky Microplate Spectrophotometer, Thermofisher Scientific).
Complementary DNA (cDNA) Synthesis and Quantification
cDNA was generated from RNA samples by reverse transcriptase PCR using M-MuLV reverse transcriptase enzyme (New England Biolab, England) according to the manufacturer’s protocol. The generated cDNA was quantified using a nanodrop spectrophotometer at 260 and 280 nm.
Serotyping of DENV
Virus identification was performed by two rounds of nested PCR using two sets of virus-specific primers (Table 2). For virus typing, the C-prM gene junction of DENV isolates was selected. The amplified product for DENV-2 was 403 base pairs (bp).

Table 2: List of primers for DENV-serotype 1-4 detection.
The reaction mixture used for the first and second rounds of PCR was defined as follows:
Master Mix: 10 µL, Outer forward primer: 1 µL, Outer reverse primer: 1 µL, PCR product: 1 µL, Nuclease free water: 7 µL, and the total volume was 20 µL.
All the reagents were well mixed on ice and mixed by vortexing them for several seconds, and then all the reaction tubes were placed in a thermal cycler (BIORAD) for amplification. The cycling temperature profile was set on a thermal cycler and the run was designed as shown below.
The cycling temperature profiles for the first and second rounds were as follows:

PCR Amplification of DENV NS3 Gene
Those samples were only selected for amplification of the NS3 gene, which gave bands for DENV serotype 2 on gel electrophoresis. The list of primers (Table 3).
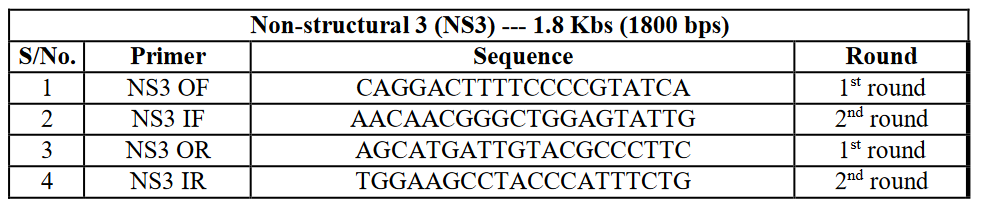
Table 3: Primer sequences for DENV NS3 gene amplification.
The protocol was optimized by performing it in triplicate with different temperatures and timings.
The reaction mixture used for the first and second rounds of PCR was defined as follows:
Master Mix: 6 µL, outer forward primer: 1 µL, outer reverse primer: 1 µL, PCR product: 4 µL, nuclease free water: 8 µL, and the total volume was 20 µL.
All the reagents were well mixed while being put on ice and mixed by vortexing for several seconds, and then all the reaction tubes were placed in a thermal cycler (BIORAD) for amplification. The cycling temperature profiles were set on the thermal cycler and the run was started as shown below:
The cycling temperature profiles for the first and second round was as follows:

The size of the NS3-amplified product was approximately 1850 bps, which is shown in (Table 4).
Gel-Electrophoresis
The PCR results were run on a 1-2% agarose gel, stained, and visualized with ethidium bromide using the Gel Documentation System (Bio-Rad, Hercules, CA, USA).
Phylogenetic Characterization
The amplified fragments were submitted to NCBI for accession numbers, and after getting the accession numbers, polygenetic analysis was done for these amplified fragments of DENV-2 to check the similarities with the DENV-2 strains detected in other areas of the world. Phylogenetic trees were made using the MEGA 11 program. And the method of analyzing phylogenetic relationships was the statistical neighbor-joining (NJ) method [22].
Cloning of the NS3-Amplified Fragment
After the amplification of the DENV NS3 gene, the amplified fragment of DENV was cloned using the TA cloning kit (Invitrogen, USA). The fragment was ligated with the TA cloning vector PCR 2.1 with 100 ng, of which the molar ratio was 1:3 of the insert to the vector. In the vector, the insert was the NS3-amplified fragment. After ligation, it was transformed into competent cells of E. coli (DH5α). The reaction mixture of the ligation was as follows: PCR 2.1 vector (25ng/ μL): 2 μL, T4 DNA Ligase (5U/ μL): 1 μL, 10 X ligation buffer: 1 μL, PCR product (~10 ng): 2 μL, sterile dH2O: 4 μL. The reaction mixture was incubated at 14°C overnight. After ligation, commercially available competent cells of E. coli bacterial cells (DH5α) were transformed with our ligated product; this we ligated first with a PCR 2.1 vector.
RESULTS
A total of 267 dengue symptomatic blood samples (156 males, 111 female) were collected from different hospitals in Khyber Pakhtunkhwa and Islamabad, Pakistan, from 2020─2022. Identification of circulating DENVs through the Immunochromatographic assay test (ICT) method was done and, out of the 267 samples 150 were dengue NS1 positive (106 males and 44 females). Of these 150 DENV NS1-positive blood samples, 89 tested positive for DENV in the first round of PCR, (54 males and 35 females), A gel image of some of the positive bands (Figure 4).
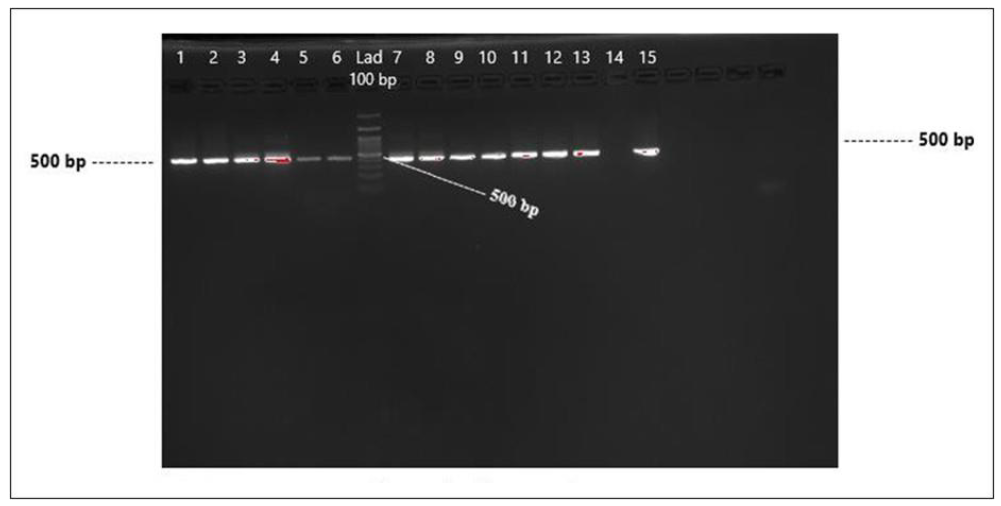
Figure 4: Detection of DENV by PCR in DENV NS1-positive serum samples (Round 1), Lanes 1-6 DENV-positive, Lane 6, 100 bp Ladder, and Lanes 7-13 and 15 DENV-positive samples.
Further PCR amplification of round 1 PCR products revealed that 48 samples were detected as DENV-2. Bands on DNA gel electrophoresis that were indicative of DENV-2 (Figure 5).
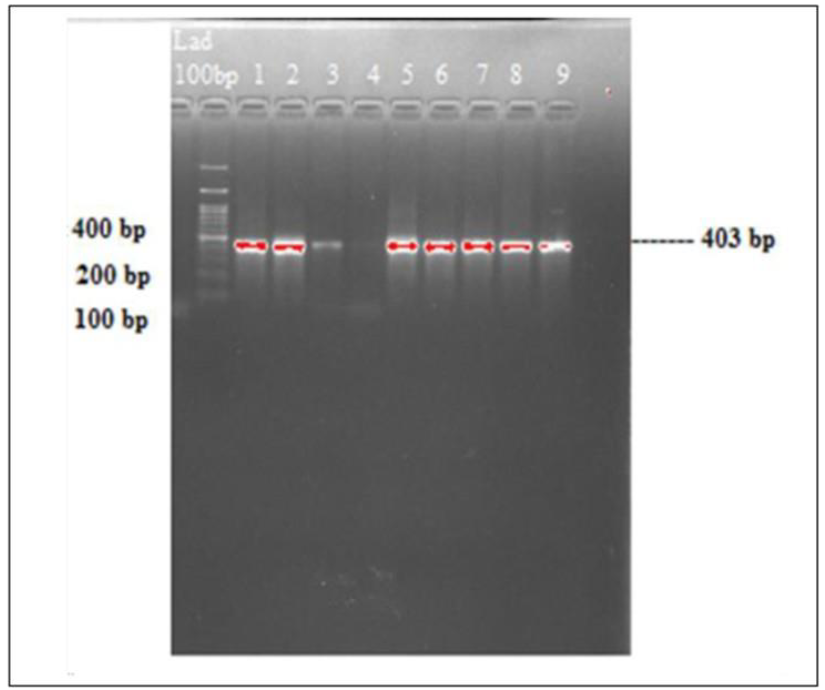
Figure 5: Gel image bands of DENV 2 virus amplification. Lane 1, 100 bp ladder and lanes 1-3, 5-9 DENV-2 bands.
Of these 48 samples, 10 samples showed bands on gel electrophoresis that were indicative of DENV-3. We concluded that there was co-infection in these patients (Figure 6).
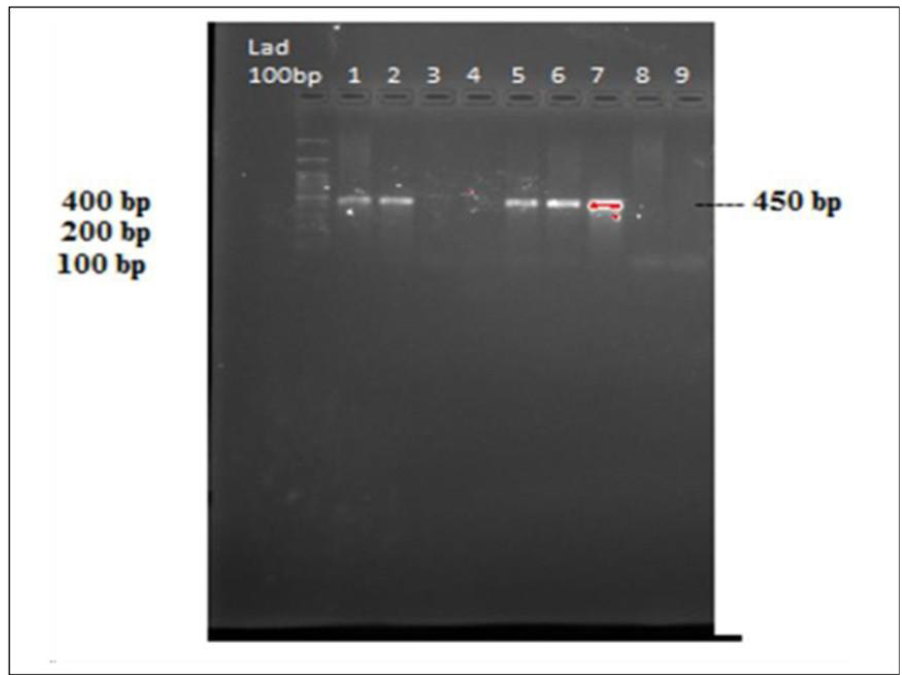
Figure 6: Gel image bands of DENV 3 virus amplification. Lane 1, 100 bp ladder and lane 1,2,5-7 DENV-3 bands.
The samples that were positive for DENV-2 only were eluted from the gel and 4 of the samples were randomly confirmed by sequencing.
Gender-Specific Cases
All 267 collected samples were checked by ICT dengue method for dengue; out of 267, 150 samples were DENV NS1-ICT-positive. In these 150 positive samples 106 were from adult males and 44 were from adult females and out of these 150 samples, 89 were PCR-positive. Out of these 89 positive samples, 54 were adult males and 35 were adult females. Therefore, we can say from above results that in Pakistan, the dengue virus can affect both males and females, but males were predominantly affected.
Phylogenetic Analysis of the DENV-2 Amplified Fragment
After the observation of amplified bands on gel electrophoresis, the bands were purified by a PCR product purification kit (GFX™ PCR DNA and Gel Band Purification Kit, Merck, KGaA, Darmstadt, Germany). Then these products were confirmed by Sanger sequencing at the Armed Forces Institute of the Bone Marrow Transplant Center, Rawalpindi, Pakistan. The accession numbers of the sequenced fragments of serotype-2 were OR797468, OR79769, OR79770, OR797471. The phylogenetic analysis was done using the Meg 11 software, and phylogenetic tree was drawn through the Neighbor joining approach of the analyzed sequences with detected strains of serotype 2 polyprotein of the dengue virus. Phylogenetic analysis showed that our detected strain of DENV-2 has shown 100 % similarity. The strain detected earlier from NIH Pakistan showed 98.68 % similarity to Chinese, Brazilian, Malaysian, Singaporean and US strains. The phylogenic tree of one the sequences accession number (OR797468) (Figure 7).

Figure 7: Phylogenetic tree of the detected strains of serotype 2 polyprotein of dengue virus.
PCR amplification of NS3 gene
After the amplification of DENV 2 and 3, the positive samples were tested by RT-PCR to amplify the NS3 gene of DENV with gene-specific primers which are listed (Table 3).
Out of these 48 DENV-2-positive samples, 25 samples were detected the NS3 gene on gel electrophoresis. The gel image of the some of the samples of amplified NS3 gene is provided (Figure 8).

Figure 8: Gel image of amplified product of DENV-2 NS3 gene.
After amplification, the amplified fragments of NS3 were cloned in a TA cloning vector (PCR 2.1) using a TA cloning kit (Invitrogen, Waltham, MA, USA). The NS3 fragment was kept for further functional analysis.
DISCUSSION
Previous studies have revealed that DENV-2 is the most common cause of dengue fever worldwide [28]. In the present study, genetic analysis of DENV-positive samples collected over a two-year period (2020-2022) were showing that DENV-2 was the most prevalent DENV in Pakistan during the COVID-19 pandemic.
Our results showed that dengue can affect both males and females, but males are predominantly affected by dengue virus, as confirmed by other studies [26,29-31]. Our results also demonstrated DENV co-infections in some of the patients. However, due to the small number of samples, the association between DENV co-infection and disease severity could not be determined as yet, and research on a larger number of samples from all over the country is underway to determine the effects of co-infection on disease severity [32]. It has been suggested that one reason for co-infection may be the co-circulation of all four serotypes in Pakistan [33]. It has also been proposed that a recombination of different DENVs may occur in co-infections which may result in more virulent and pathogenic strains [31]. Some former studies found that coinfection with two or multiple serotypes or genotypes may affect the disease representation [34-36]. Previous studies suggested that DENV co-infection can also occur within mosquitoes (Aedes aegypti), due to multiple blood meals in mosquitoes during their gonotrophic cycle [37].
Our research also focuses on the NS3 gene, which is a non-structural gene of 1.8 kb size involved in RNA synthesis and DENV genome replication. The NS3 protein consists of multiple catalytic domains, that perform different functions like helicase, nucleoside triphosphatase, and NTPase activities during RNA synthesis. Viral RNA replication also requires a portion of 40 residues from the NS2B gene as its cofactor for NS3 protease activity. The helicase/ATPase domain is located at the C-terminus of the NS3 gene [38]. NS3 is, therefore, one of the major targets for the development of antiviral drugs against dengue viral infection.
The phylogenetic analysis of four randomly sequenced samples from 48 positive DENV-2 samples showed a cosmopolitan genotype. The detected strain of DENV-2 shows above 99% similarity with the viruses detected from India, China, Singapore, Malaysia, and Brazil from 2019 to 2023, which might be explained by an ongoing virus exchange between these countries. They are among the aviation hubs with the greatest global connectivity with increasing transport of people with a direct impact on the inbound and outgoing transmission of dengue viruses [20,39].
Phylogenetic analyses of the envelope, pre-membrane (prM), and capsid (C) genes show that all DENV-2 isolates from Pakistan collected since 2007 belong to a single monophyletic branch. These viruses belong to the cosmopolitan genotype IV group and are most closely related to viruses found in China, India, and Sri Lanka in the previous 20 years [40,41].
To control the fast-growing number of dengue cases, there is an urgent need for new therapeutic approaches. Currently, several antiviral drugs are tested in pre-clinical and clinical trials. However, there is still no clinically approved treatment available for DENV infection. In this study, we analyzed the circulating DENV serotypes in Pakistan during the 2020-2022 outbreaks and observed that DENV-2 was the predominantly circulating DENV. However, co-infections with DENV-3 were also observed in some samples. The presence of dengue co-infections is of concern as they might lead to more severe infections and a high morbidity and mortality rate since Pakistan was endemic for dengue for the last 3 decades. We were also able to amplify and obtain clones of the DENV NS3 gene in this study. In the future, we aim to express the NS3 gene in mammalian cells and check for inhibition of the NS3 gene through RNAi and antiviral drugs, which may help to develop targeted therapeutics against DENV infection. This study highlights the importance of proper monitoring and surveillance of circulating serotypes of the DENV during outbreaks, which will aid in early diagnostic and disease control measures.
CONCLUSION
The present study from Pakistan, discovered the most prevalent Dengue serotype from a period from 2020 to 2022, serotype-2, and co-infection with serotype-3 in some cases, and showed that males were more affected than females. In Pakistan, co-infection with distinct genotypes or serotypes may increase the risk and severity of the disease in later outbreaks. This study also characterized the DENV-NS3 gene which will be cloned in a TA-cloning vector and will be used for further functional analysis of dengue viruses, which will aid in the development of precise and early diagnostics and treatment.
CONFLICT OF INTEREST
There is no conflict of interest to declare.
ACKNOWLEDGMENT AND FUNDING INFORMATION
We thank several diagnostic laboratories and clinicians for providing blood samples. This study was supported by a HEC (Higher Education Commission of Pakistan) funded project NRPU-8403 at National University of Medical Sciences, Rawalpindi, Pakistan and partially supported by the Research Center for Precision Environmental Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan and by Kaohsiung Medical University Research Center Grant (KMU-TC113A01).
AUTHOR CONTRIBUTIONS
Muhammad Bilal Khan performed the experimental work and wrote the manuscript. Rehana Rani and Deeba Amraiz assisted with manuscript writing. Yeou-Lih Huang and Hans-Uwe Dahms assisted with the review and editing, and Liaqat Ali supervised all the experimental work, resources, funding acquisition, and project administration. All the authors read and approved the submitted manuscript version here.
REFERENCES
- M. Yousaf, A. Siddique, U. Ashfaq, Muhammad Ali. Scenario of dengue infection & its control in Pakistan: An up—date and way forward. Asian Pac J Trop Med. 2018;11(1):241-247.
- Dash P, Parida M, Saxena P, Abhyankar A, Singh C, Tewari K, et al. Reemergence of dengue virus type-3 (subtype-III) in India: Implications for increased incidence of DHF & DSS. Virol J. 2006;3:55.
- Sherin A. Dengue Fever: A major public health concern in Pakistan. Khyber Medical University Journal. 2011;3(1):1-3.
- Shaikh OA, Baig MT, Tahir S, Parekh AE, Nashwan AJ. Dengue outbreak following unprecedented flooding in Pakistan. Hyg Environ Health Adv. 2023;7:100076.
- https://www.nature.com/scitable/topicpage/dengue-viruses-22400925/
- Schmidt AG, Lee K, Yang PL, Harrison SC. Small-molecule inhibitors of dengue-virus entry. PLoS Pathog. 2012;8(4):e1002627.
- Yousaf MZ, Siddique A, Ashfaq U, Ali M. Scenario of dengue infection & its control in Pakistan: An up-date and way forward. Asian Pac J Trop Med. 2018;11(1):15.
- Parkash O, Shueb RH. Diagnosis of Dengue Infection Using Conventional and Biosensor Based Techniques. Viruses. 2015;7(10):5410-5427.
- Ahmad A, Aziz MA, Aftab A, Ullah Z, Ahmad MI, Mustan A. Epidemiology of dengue in Pakistan, present prevalence and guidelines for future control. Int J Mosq Res. 2017;4(6):25-32.
- Khattak A, Khan S, Ali I, Gul A, Khabir MN, Javed B. Burden and distribution of dengue infection in Pakistan (2000-19): a review. Braz J Biol. 2023;:84:e267982.
- Gubler DJ. Dengue and Dengue Hemorrhagic Fever. Clin Microbiol Rev. 1998;11(3):480–496.
- World Health Organization. Regional Office for the Western, P., Dengue - Pakistan. 2022.(https://iris.who.int/handle/10665/352792).
- Dengue, W., Guidelines for diagnosis, Treatment. Prevention and Control. (No Title), 2009.(https://www.who.int/publications/i/item/9789241547871).
- Kuczera D, Assolini JP, Tomiotto-Pellissier F, Pavanelli WR, Silveira GF. Highlights for Dengue Immunopathogenesis: Antibody-Dependent Enhancement, Cytokine Storm, and Beyond. J Interferon Cytokine Res. 2018;38(2):69-80.
- W.H.O. Comprehensive guideline for prevention and control of dengue and dengue haemorrhagic fever. 2011.(https://iris.who.int/handle/10665/204894)
- Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol. 2010;84(20):10438-10447.
- Fatima Z, Idrees M, Bajwa MA, Tahir Z, Ullah O, Zia MQ, et al. Serotype and genotype analysis of dengue virus by sequencing followed by phylogenetic analysis using samples from three mini outbreaks-2007-2009 in Pakistan. BMC Microbiol. 2011;11:200.
- Khan MB, Yang ZS, Lin CY, Hsu MC, Urbina AN, Assavalapsakul W, et al. Dengue overview: An updated systemic review. J Infect Public Health. 2023;16(10):1625-1642.
- Qureshi JA, Notta NJ, Salahuddin N, Zaman V, Khan JA. An epidemic of Dengue fever in Karachi--associated clinical manifestations. J Pak Med Assoc. 1997;47(7):178-81.
- Umair M, Haider SA, Rehman Z, Jamal Z, Ali Q, Hakim R, et al. Genomic Characterization of Dengue Virus Outbreak in 2022 from Pakistan. Vaccines (Basel). 2023;11(1):163.
- Cobelens FG, Groen J, Osterhaus AD, Leentvaar-Kuipers A, Wertheim-van Dillen PM, Kager PA. Incidence and risk factors of probable dengue virus infection among Dutch travellers to Asia. Trop Med Int Health. 2002;7(4):331-8.
- Paul RE, Patel AY, Mirza S, Fisher-Hoch SP, Luby SP. Expansion of epidemic dengue viral infections to Pakistan. Int J Infect Dis. 1998;2(4):197-201.
- Khanani MR, Arif A, Shaikh R. Dengue in Pakistan: Journey from a disease free to a hyper endemic nation. J Dow Univ Health Sci. 2011;5(3):81-84.
- Suleman M, Faryal R, Alam MM, Khurshid A, Sharif S, Shaukat S, et al. Outbreak of dengue virus type-3 in Malakand, Pakistan 2015; A laboratory perspective. Acta Trop. 2017;169:202-206.
- Akram D, Igarashi A, Takasu T. Dengue virus infection among children with undifferentiated fever in Karachi. Indian J Pediatr. 1998;65:735-740.
- Khan J, Adil M, Wang G, Tsheten T, Zhang D, Pan W, et al. A cross-sectional study to assess the epidemiological situation and associated risk factors of dengue fever; knowledge, attitudes, and practices about dengue prevention in Khyber Pakhtunkhwa Province, Pakistan. Front Public Health. 2022;10:923277.
- Abid MA, Abid MB. Climate change and the increased burden of dengue fever in Pakistan. Lancet Infect Dis. 2023;23(1):17-18.
- World Health Organization. Regional Office for the Western, P., Dengue Situation Updates 2021. 2021, WHO Regional Office for the Western Pacific: Manila.(https://iris.who.int/handle/10665/341149?show=full)
- Mehmood A, Khalid Khan F, Chaudhry A, Hussain Z, Laghari MA, Shah I, et al. Risk Factors Associated with a Dengue Fever Outbreak in Islamabad, Pakistan: Case-Control Study. JMIR Public Health Surveill . 2021;7(12):e27266.
- Khan J, Khan I, Ghaffar A, Khalid B. Epidemiological trends and risk factors associated with dengue disease in Pakistan (1980-2014): a systematic literature search and analysis. BMC Public Health. 2018;18(1):745.
- Hanan F, Ahmad J, Hadi S, Ali I, Fahim M, Qayum A. Analysis of Dengue Virus Genotypes and Further Investigations for Mixed Infections through RT-PCR in Classical Dengue Fever Patients in Pakistan. Int J Pathol. 2022;72-78.
- Bharaj P, Chahar HS, Pandey A, Diddi K, Dar L, Guleria R, et al. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol J. 2008;5:1.
- Worobey M, Rambaut A, Holmes EC. Widespread intra-serotype recombination in natural populations of dengue virus. Proc Natl Acad Sci USA. 1999;96(13):7352-7357.
- Dhanoa A, Hassan SS, Ngim CF, Lau CF, Chan TS, Adnan NA, et al. Impact of dengue virus (DENV) co-infection on clinical manifestations, disease severity and laboratory parameters. BMC Infect Dis. 2016;16(1):406.
- Senaratne UTN, Murugananthan K, Sirisena PDNN, Carr JM, Noordeen F. Dengue virus co-infections with multiple serotypes do not result in a different clinical outcome compared to mono-infections. Epidemiol Infect. 2020;148:e119.
- Colombo TE, Vedovello D, Mondini A, Reis AFN, Cury AAF, Oliveira F, et al. Co-infection of Dengue virus by serotype 1 and 4 in patients from a medium sized city from Brazil. Rev Inst Med Trop Sao Paulo. 2013;55.
- Tazeen A, Afreen N, Abdullah M, Deeba F, Haider SH, Kazim SN, et al. Occurrence of co-infection with dengue viruses during 2014 in New Delhi, India. Epidemiol Infect. 2017;145(1):67-77.
- Luo D, Xu T, Hunke C, Grüber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82(1):173-183.
- Yenamandra SP, Koo C, Chiang S, Lim HS, Yeo ZY, Ng LC, et al. Evolution, heterogeneity and global dispersal of cosmopolitan genotype of Dengue virus type 2. Sci Rep. 2021;11:13496.
- Khan MA, Ellis EM, Tissera HA, Alvi MY, Rahman FF, Masud F, Emergence and diversification of dengue 2 cosmopolitan genotype in Pakistan, 2011. PLoS One. 2013;8(3):e56391.
- Koo C, Nasir A, Hapuarachchi HC, Lee KS, Hasan Z, Ng LC, et al. Evolution and heterogeneity of multiple serotypes of Dengue virus in Pakistan, 2006-2011. Virol J. 2013;10:275.